cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
Por um escritor misterioso
Descrição
lt;p>Single-cell RNA sequencing reveals the gene structure and gene expression status of a single cell, which can reflect the heterogeneity between cells. However, batch effects caused by non-biological factors may hinder data integration and downstream analysis. Although the batch effect can be evaluated by visualizing the data, which actually is subjective and inaccurate. In this work, we propose a quantitative method cKBET, which considers the batch and cell type information simultaneously. The cKBET method accesses batch effects by comparing the global and local fraction of cells of different batches in different cell types. We verify the performance of our cKBET method on simulated and real biological data sets. The experimental results show that our cKBET method is superior to existing methods in most cases. In general, our cKBET method can detect batch effect with either balanced or unbalanced cell types, and thus evaluate batch correction methods.</p>

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
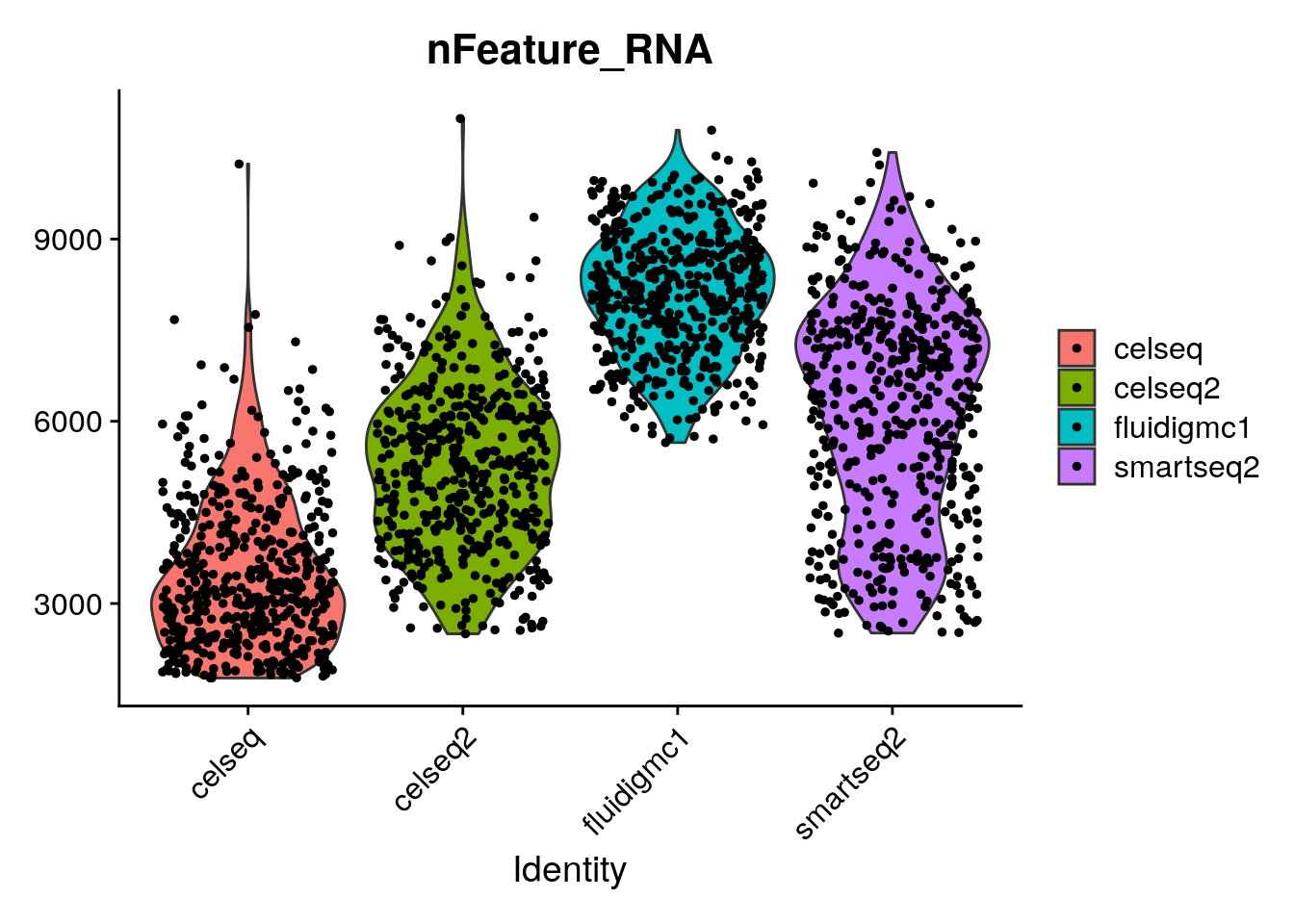
12 Batch Correction Lab ANALYSIS OF SINGLE CELL RNA-SEQ DATA
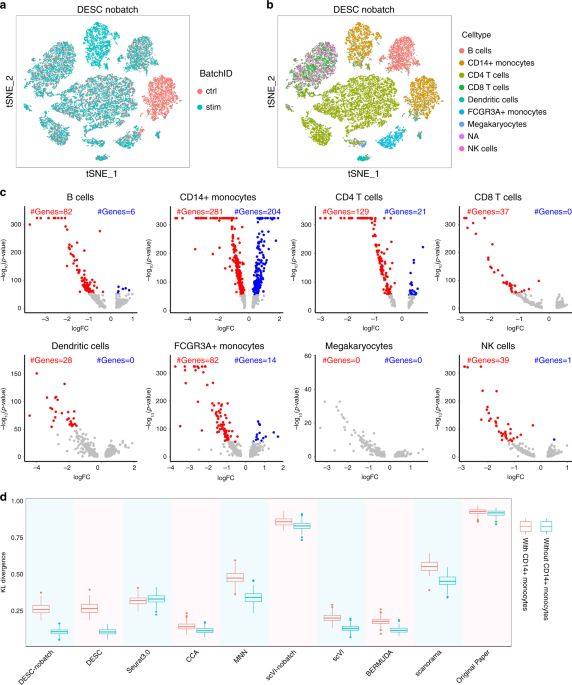
Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis

Assessment of batch-correction methods for scRNA-seq data with a new test metric
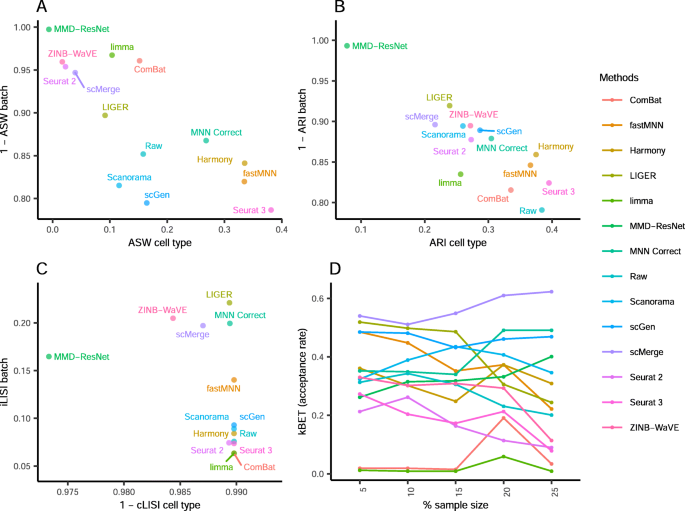
A benchmark of batch-effect correction methods for single-cell RNA sequencing data, Genome Biology
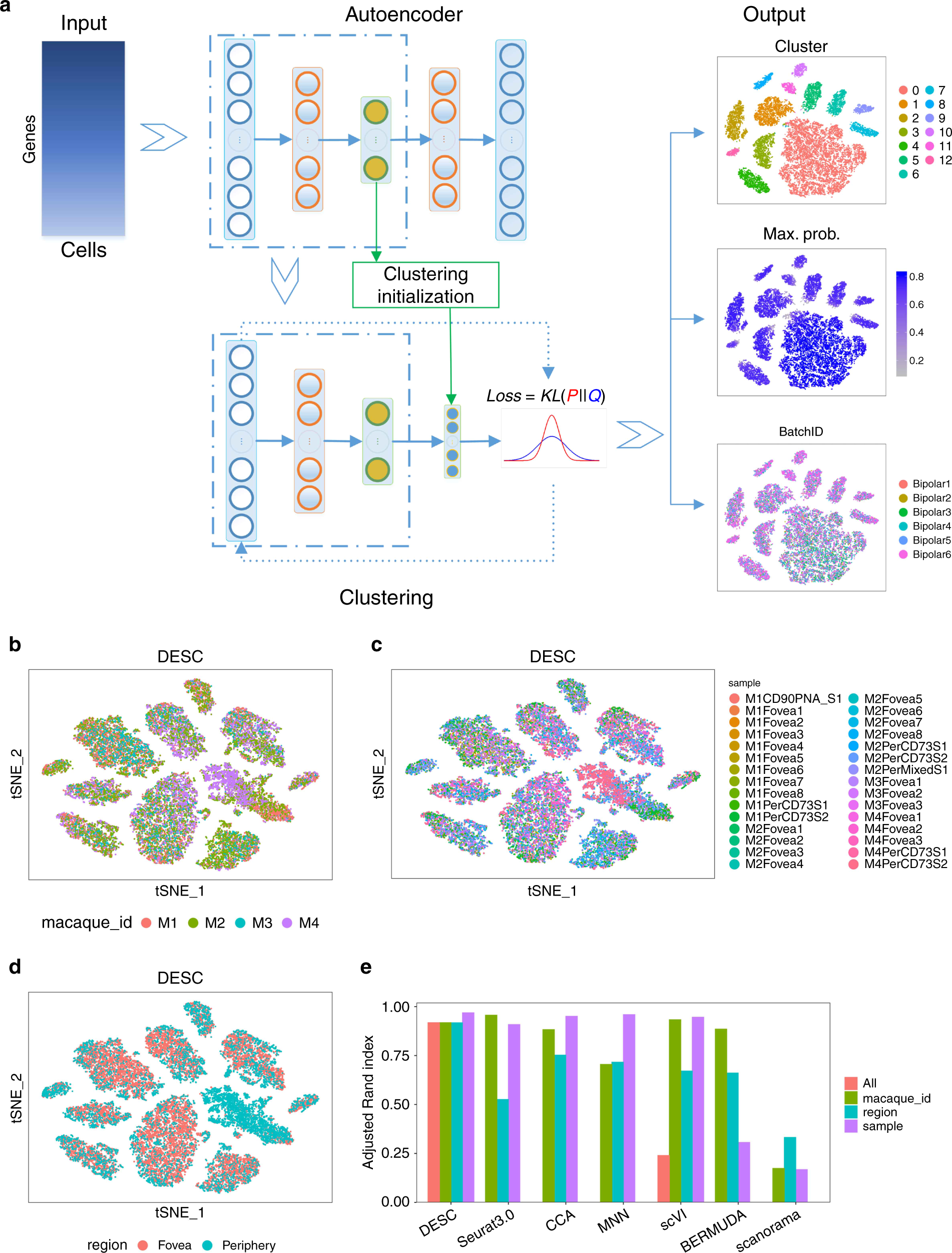
Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis
PDF) Comparison of Scanpy-based algorithms to remove the batch effect from single-cell RNA-seq data

How to Batch Correct Single Cell. Comparing batch correction methods for…, by Nikolay Oskolkov

PDF) BEENE: Deep Learning based Nonlinear Embedding Improves Batch Effect Estimation

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
de
por adulto (o preço varia de acordo com o tamanho do grupo)






