Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Descrição

Deep learning-based molecular dynamics simulation for structure-based drug design against SARS-CoV-2 - ScienceDirect
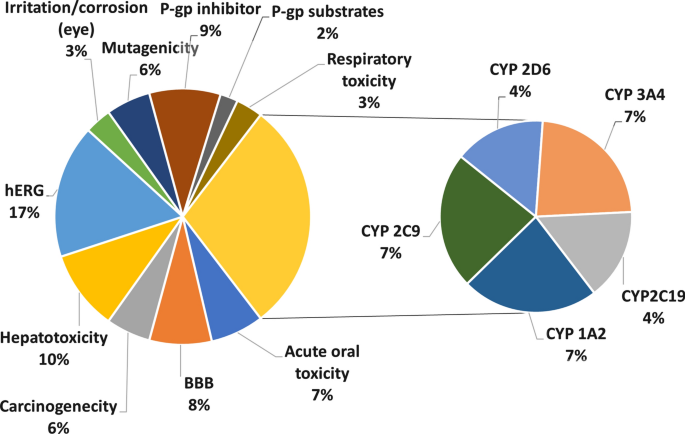
Machine learning models for classification tasks related to drug safety

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
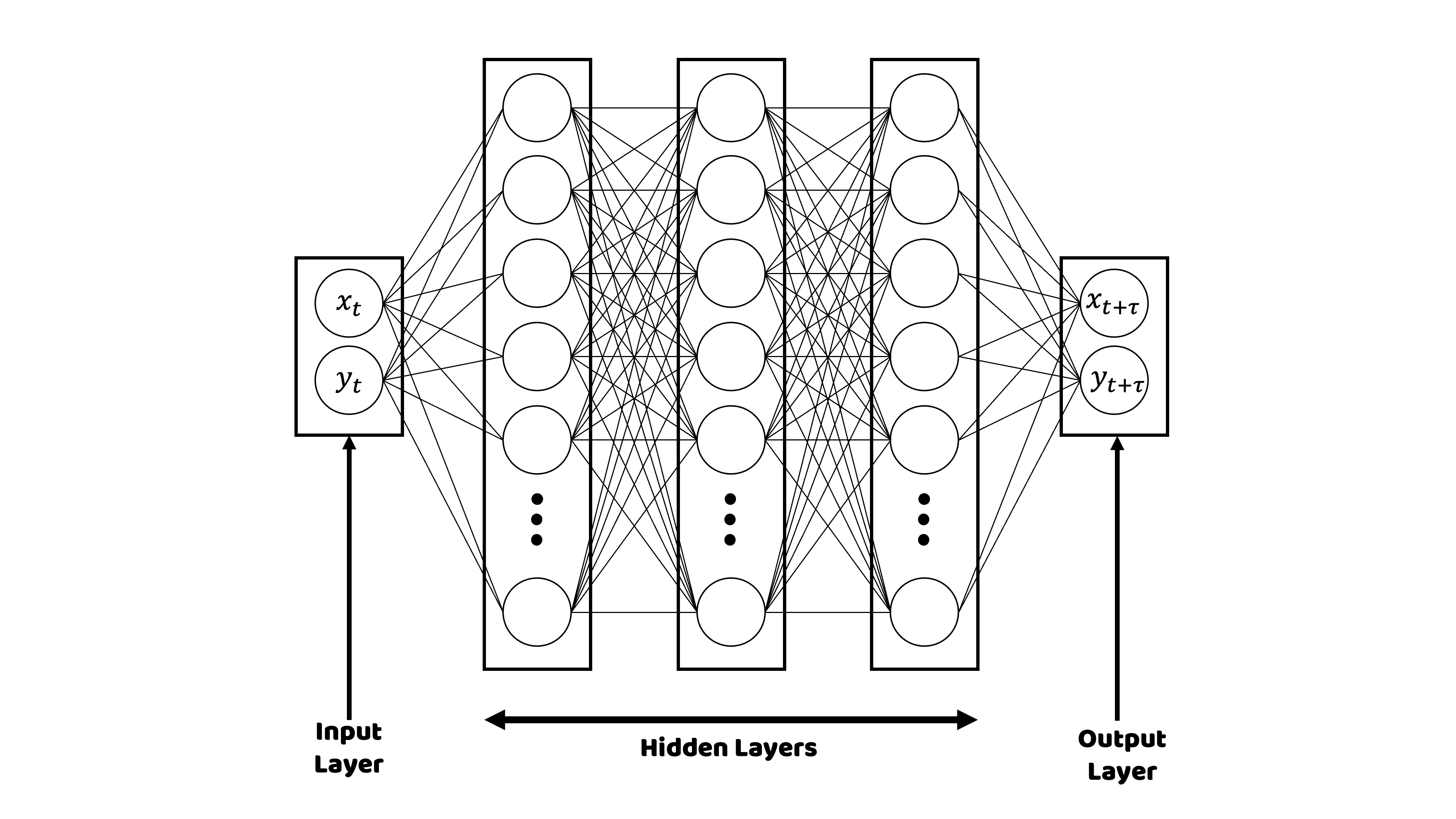
BioSimLab - Research

P-glycoprotein Substrate Models Using Support Vector Machines Based on a Comprehensive Data set

Fast fit (A) and best fit (B) to the Catalyst common features model for
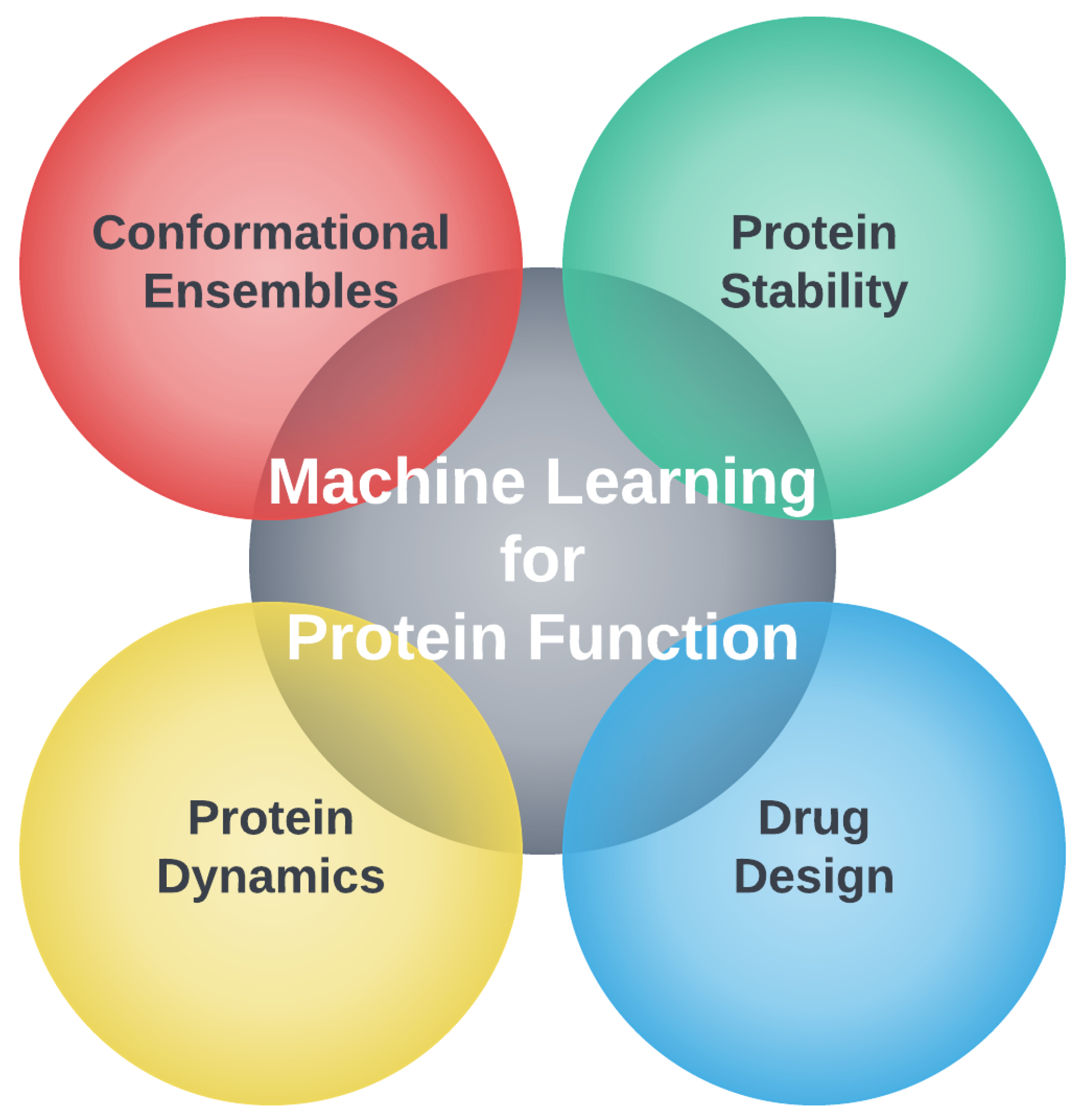
Biomolecules, Free Full-Text
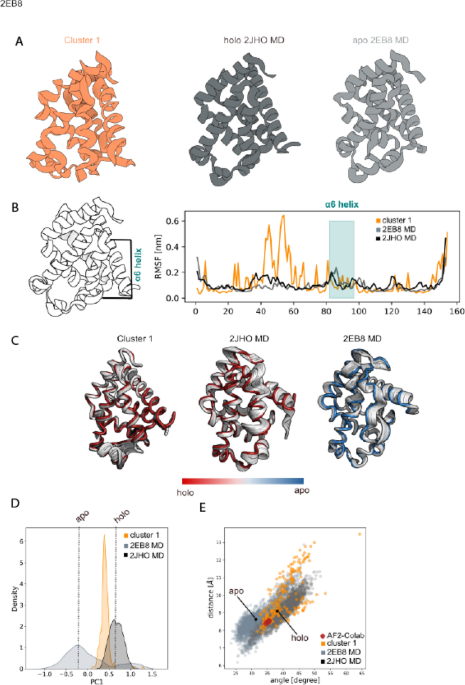
Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble

Structures of P-glycoprotein in different conformations (A) Domain

Molecular modeling of human P-gp structure. (a) 3D Structure of human
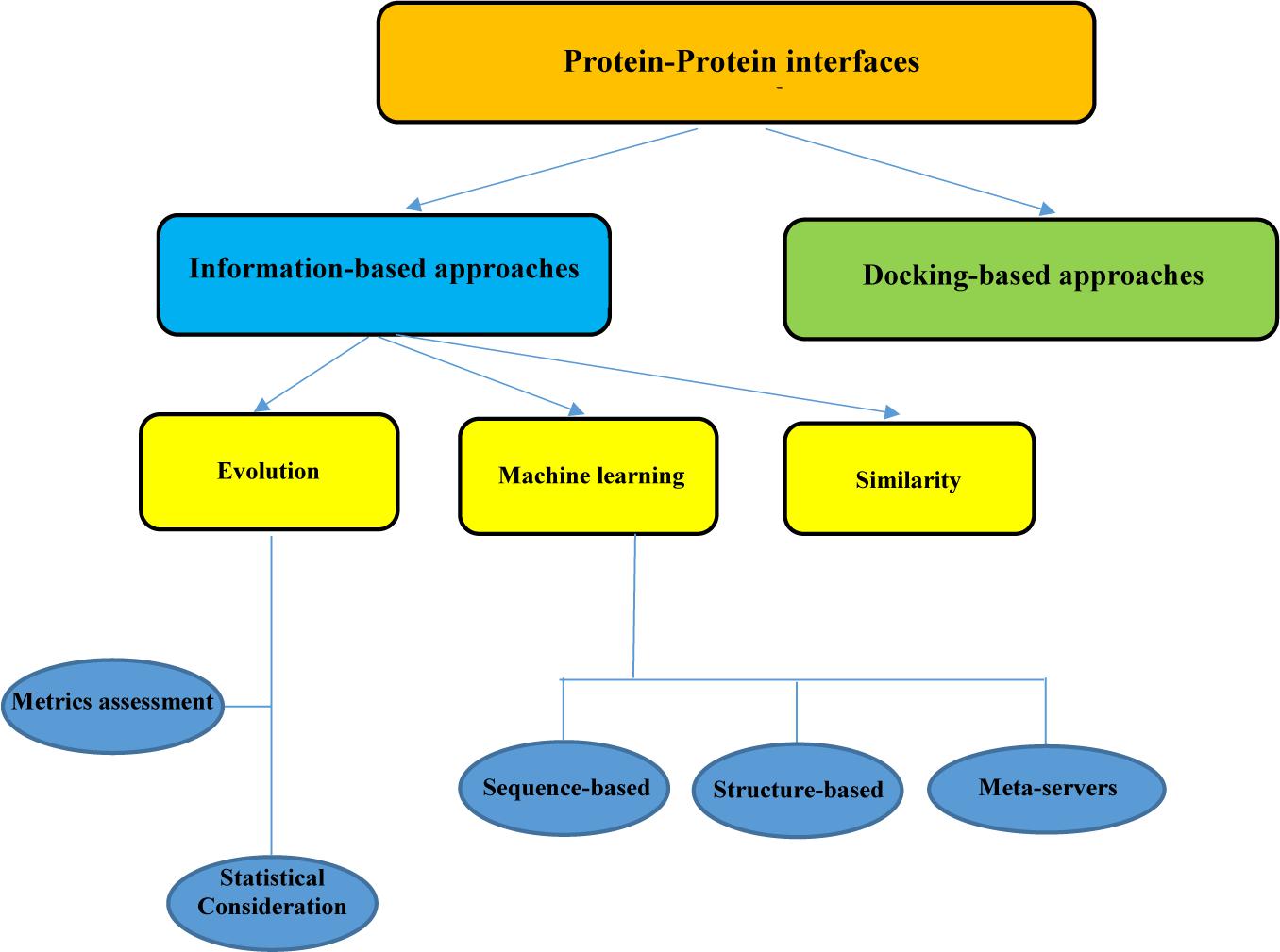
Frontiers In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions
de
por adulto (o preço varia de acordo com o tamanho do grupo)