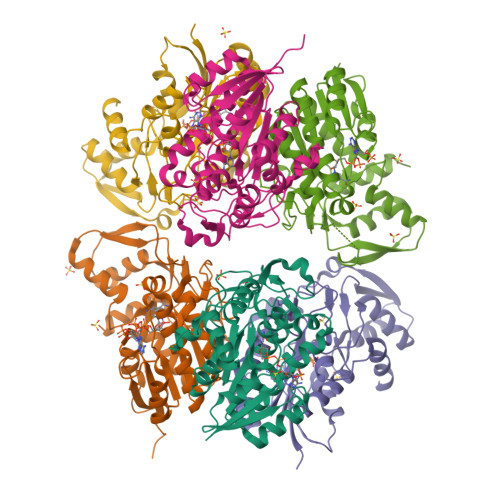
RCSB PDB - 4H6R: Structure of reduced Deinococcus radiodurans
Por um escritor misterioso
Descrição
Structure of reduced Deinococcus radiodurans proline dehydrogenase
RCSB PDB - 3SC6: 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP

RCSB PDB - 4H6Q: Structure of oxidized Deinococcus radiodurans proline dehydrogenase complexed with L-tetrahydrofuroic acid

RCSB PDB - 6JP Ligand Summary Page
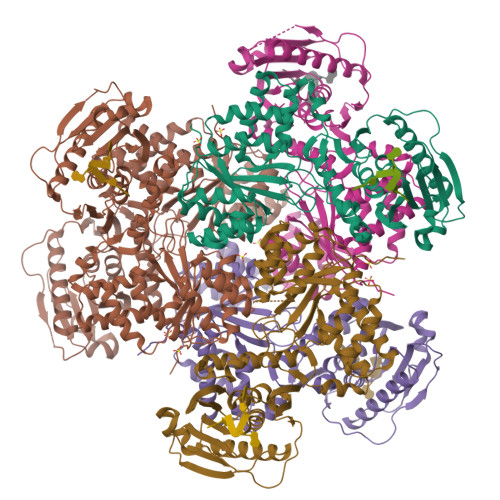
RCSB PDB - 6VM6: Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 2'3'3'-cAAA
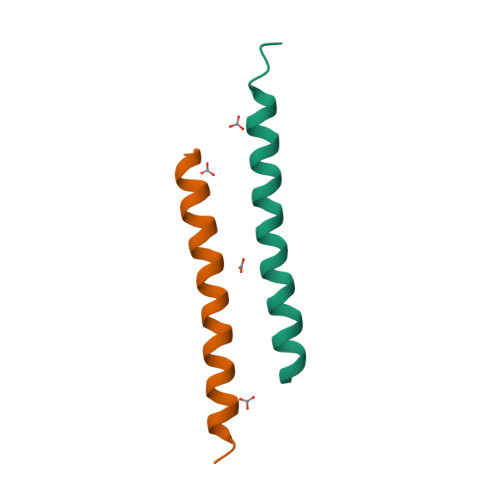
RCSB PDB - 5NNM: The crystal structure of dimeric LL-37
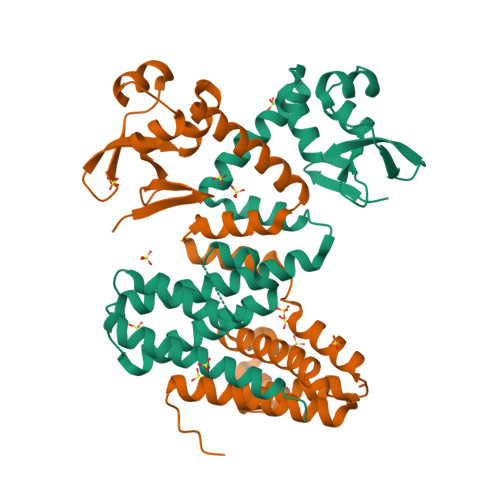
RCSB PDB - 7QVB: Crystal structure of the DNA-binding protein DdrC from Deinococcus radiodurans

RCSB PDB - 6YRU: Crystal structure of FAP in the dark at 100K

The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound - ScienceDirect

Surface (S)-layer proteins of Deinococcus radiodurans and their utility as vehicles for surface localization of functional proteins - ScienceDirect
PDF) A covalent adduct of MbtN, an acyl-ACP dehydrogenase from Mycobacterium tuberculosis, reveals an unusual acyl-binding pocket

RCSB PDB - 4QI7: Cellobiose dehydrogenase from Neurospora crassa, NcCDH
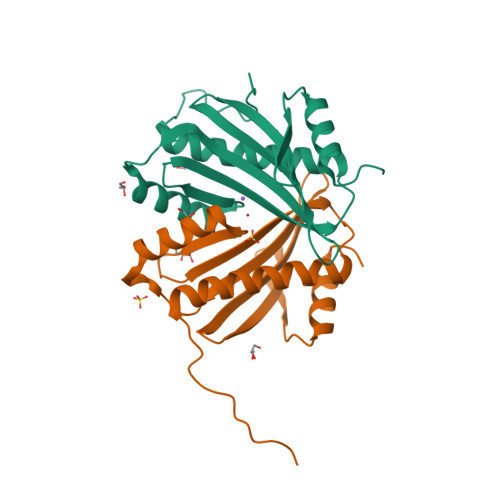
RCSB PDB - 4H5B: Crystal Structure of DR_1245 from Deinococcus radiodurans
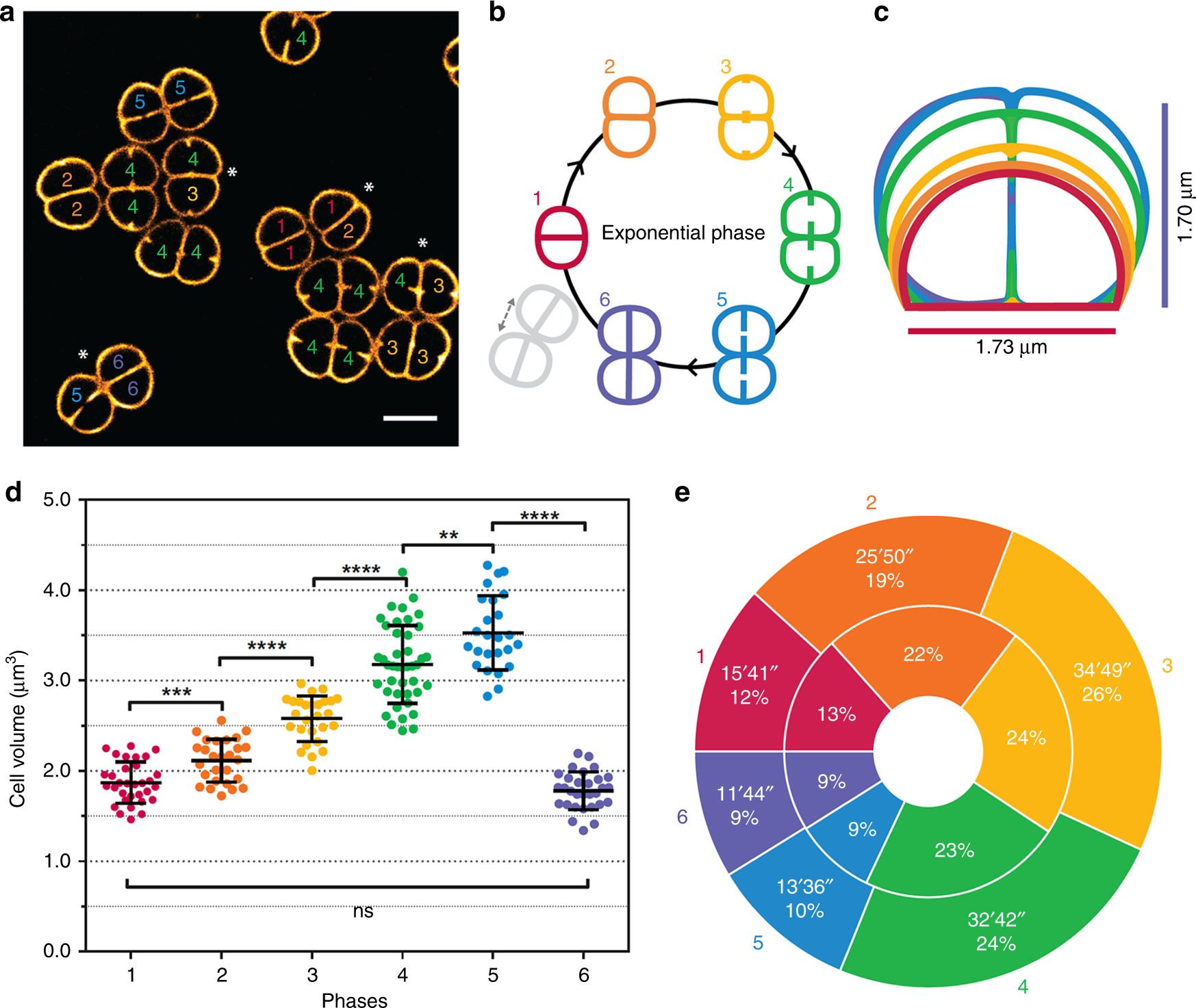
Cell morphology and nucleoid dynamics in dividing Deinococcus radiodurans

RCSB PDB - 6U9D: Saccharomyces cerevisiae acetohydroxyacid synthase

RCSB PDB - 4AR4: Neutron crystallographic structure of the reduced form perdeuterated Pyrococcus furiosus rubredoxin to 1.38 Angstrom resolution.
de
por adulto (o preço varia de acordo com o tamanho do grupo)